Cecile Breyton, IBS : How do siphophages perforate the bacterial envelope?
Background:
Phages, bacterial viruses, are fascinating nanomachines that infect very specifically bacterial hosts. They represent the most abundant biological entities on earth, and their study is getting increasingly popular in fields as diverse as ecology, genetics, phylogeny, nanophysics, agriculture and medicine. The vast majority of phages bear a protein capsid con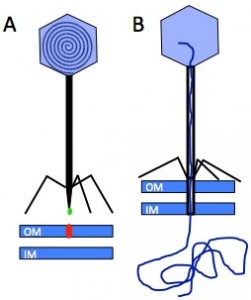 taining a densely packed dsDNA, and a tail, whose adsorption device at its end, the tail tip, serves to recognise the host surface and triggers the delivery of the phage genome into the bacterium’s cytoplasm. Infection of E. coli by the siphophage T5 is initiated by the irreversible binding of T5 tail tip to the outer membrane transporter FhuA (Fig. 1). This signal is transmitted along the tail allowing the opening of the capsid, and induces conformation changes within the tail tip that results in the perforation of the host cell wall.
taining a densely packed dsDNA, and a tail, whose adsorption device at its end, the tail tip, serves to recognise the host surface and triggers the delivery of the phage genome into the bacterium’s cytoplasm. Infection of E. coli by the siphophage T5 is initiated by the irreversible binding of T5 tail tip to the outer membrane transporter FhuA (Fig. 1). This signal is transmitted along the tail allowing the opening of the capsid, and induces conformation changes within the tail tip that results in the perforation of the host cell wall.
Figure 1: Interaction between the Receptor Binding Protein (green) of phage T5 and its host receptor, the outer membrane (OM) transporter FhuA (red) (IM: inner membrane) (A), allows capsid opening and perforation of the cell wall to channel the DNA into the host cytoplasm (B).
Objectives:
The project aims to investigate i) the structure of the tail and of the host-adsorption/perforation device composed of 5 proteins within the phage tail, ii) that of the DNA channel once the tail inserted into the cell envelope, and iii) the mechanism creating the DNA channel.
Team:
IBS: Cécile Breyton, Guy Schoehn
BIG: Wim Burmeister
GRAL recruitment: Charles Arnaud, PhD student (2014-2017)
Research plan
In an integrated approach, we will work at different levels of resolution: i) at the level of biochemical and immuno-localisation, to identify the protein that form the channel; ii) at the structural level solving the atomic structures of tail proteins and subcomplexes thereof. iii) on a more integrated scale where we will determine, by cryo-electron microscopy, the structure of the host-absorption complex at different stages of infection, i.e. before and after interaction with FhuA, either solubilised in detergent or reconstituted into liposomes and/or nanodiscs, or in vivo using E. coli mini cell.
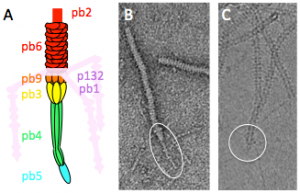
We have already identified and localised all the proteins of the tail (Zivanovic et al., J. Virol, 2014), characterised the Receptor Binding Protein-FhuA complex (Flayhan et al., Biochimie, 2012; Breyton et al., JBC, 2013), and solved the atomic structure of the distal tail protein (Flayhan et al., J. Virol, 2014). We can purify functional tails, which will be an interesting tool in biochemistry and cryo-EM due to the absence of the capsid and ejected DNA (Fig. 2).
Originality and expected results
The mechanism of bacterial cell wall perforation by phages has remained a complete mystery until quite recently. Rossmann et al., have unravelled part of this mechanism (syringe-like) for myophage T4 (long contractile tail) (Kanamura et al., Nature, 2002; Leiman et al., Cell, 2004). Its elucidation for siphophages (long flexible tail, 60% of phages), thought to be different, will provide new insights into the various mechanisms used to perforate the bacterial envelop. This study is of main interest for applications of phage therapy in medicine, but also to understand the mechanism of the siphophage-related F-pyocines, bacteriocins secreted by 90% of P. aeruginosa strains to kill competitor stains.
Figure 2: Phage T5 tail tip. A. Schematic representation. The tail tip complex is composed of pb9, pb3, pb4, pb5 and pb2. Pb1 and p132 form the L-shaped fibres, which reversibly bind the lipopolysaccharide. B. and C. purified T5 tails in electron microscopy (EM) without (B, negative stain) or with incubation with FhuA (C, cryo). The tail tip is circled. Diameter of the tail: 10 nm.
Publications:
- Noirclerc-Savoye M, Flayhan A, Pereira C, Gallet B, Gans P, Ebel C, Breyton C. (2015) Tail proteins of phage T5: investigation of the effect of the His-Tag position, from expression to crystallisation. Protein Expr Purif. 109:70-8
- Flayhan A, Vellieux FMD, Lurz R, Maury O, Contreras-Martel C, Girard E, Boulanger P & Breyton C. (2014) Crystal structure of pb9, the distal tail protein of bacteriophage T5: A conserved structural motif among all siphophages. J Virol. 88:820-8
- Zivanovic Y, Confalonieri L, Ponchon L, Lurz R, Chami M, Flayhan A, Renouard M, Huet A, Conway JF, Decottignies P, Davidson AR, Breyton C, Boulanger P. (2014) Insights into bacteriophage T5 structure and assembly from the morphogenesis genes and proteome analysis. Virol, 88, 1162-74
- Breyton C, Flayhan A, Gabel F, Lethier M, Durand G, Boulanger P, Chami M and Ebel C. (2013) Assessing the conformation changes of pb5, the Receptor Binding Protein of phage T5, upon binding to its coli receptor FhuA. J. Biol. Chem. 288:30763-72